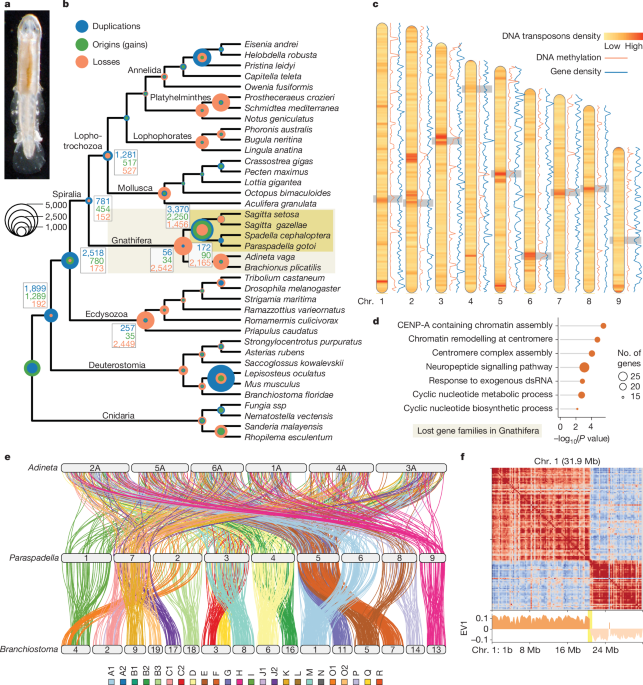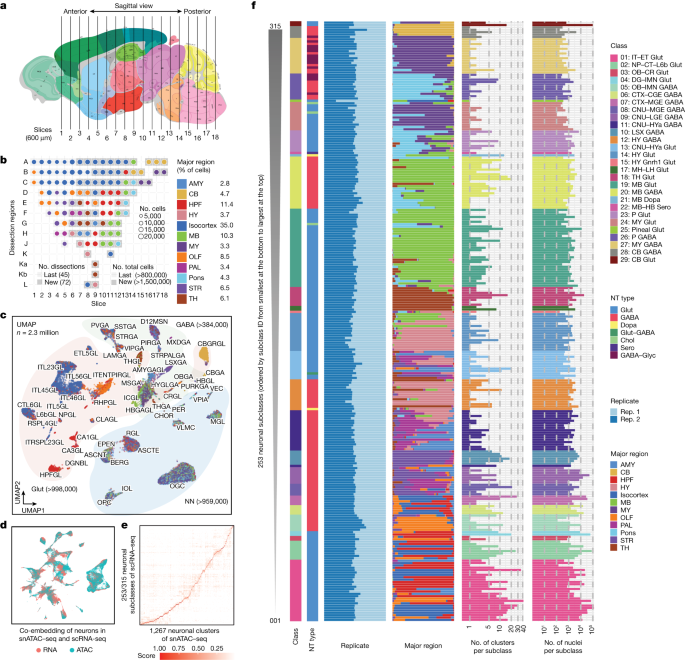
Comprehensive Single-Cell Atlas Maps Arabidopsis Life Cycle
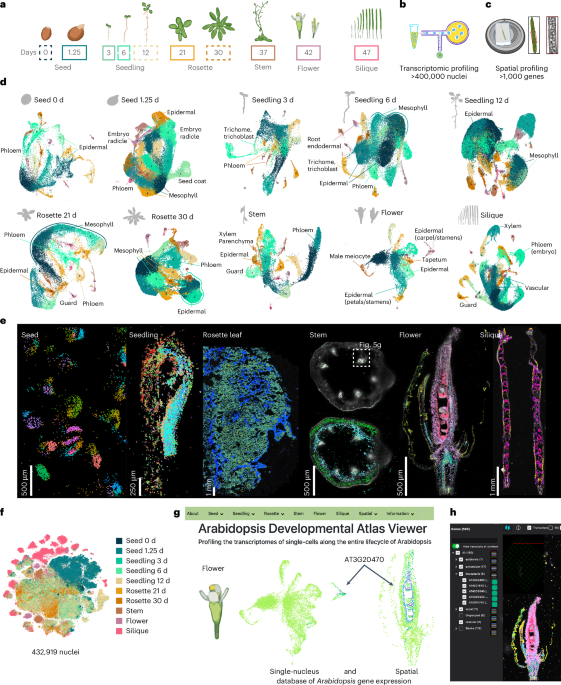
This study presents a comprehensive single-nucleus and spatial transcriptomic atlas of the Arabidopsis plant life cycle, revealing diverse molecular identities, cell types, and states across various organs and developmental stages, and demonstrating the power of combined technologies to uncover cellular complexity and function in plant biology.