AI Advances in Antibody Design Accelerate Drug Development and Viral Defense
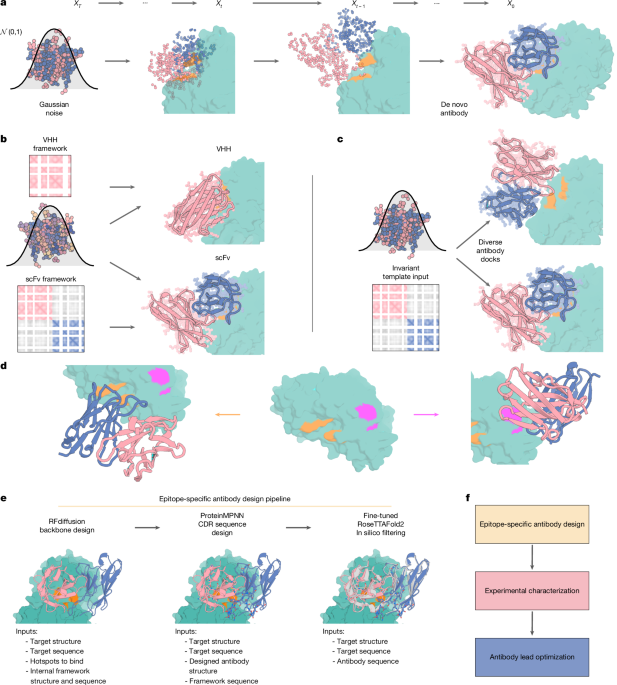
The article discusses a novel computational approach using RFdiffusion, fine-tuned on antibody structures, to design de novo antibodies and VHHs with atomic-level accuracy, targeting specific epitopes. The method enables the creation of antibodies with precise epitope targeting, validated by cryo-EM structures, and demonstrates potential for rapid therapeutic development, although current success rates are low and can be improved with advanced filtering techniques like AlphaFold3.