"Revolutionizing Avian Evolution: Computational Tools and AI Algorithm Unveil New Bird Family Tree"
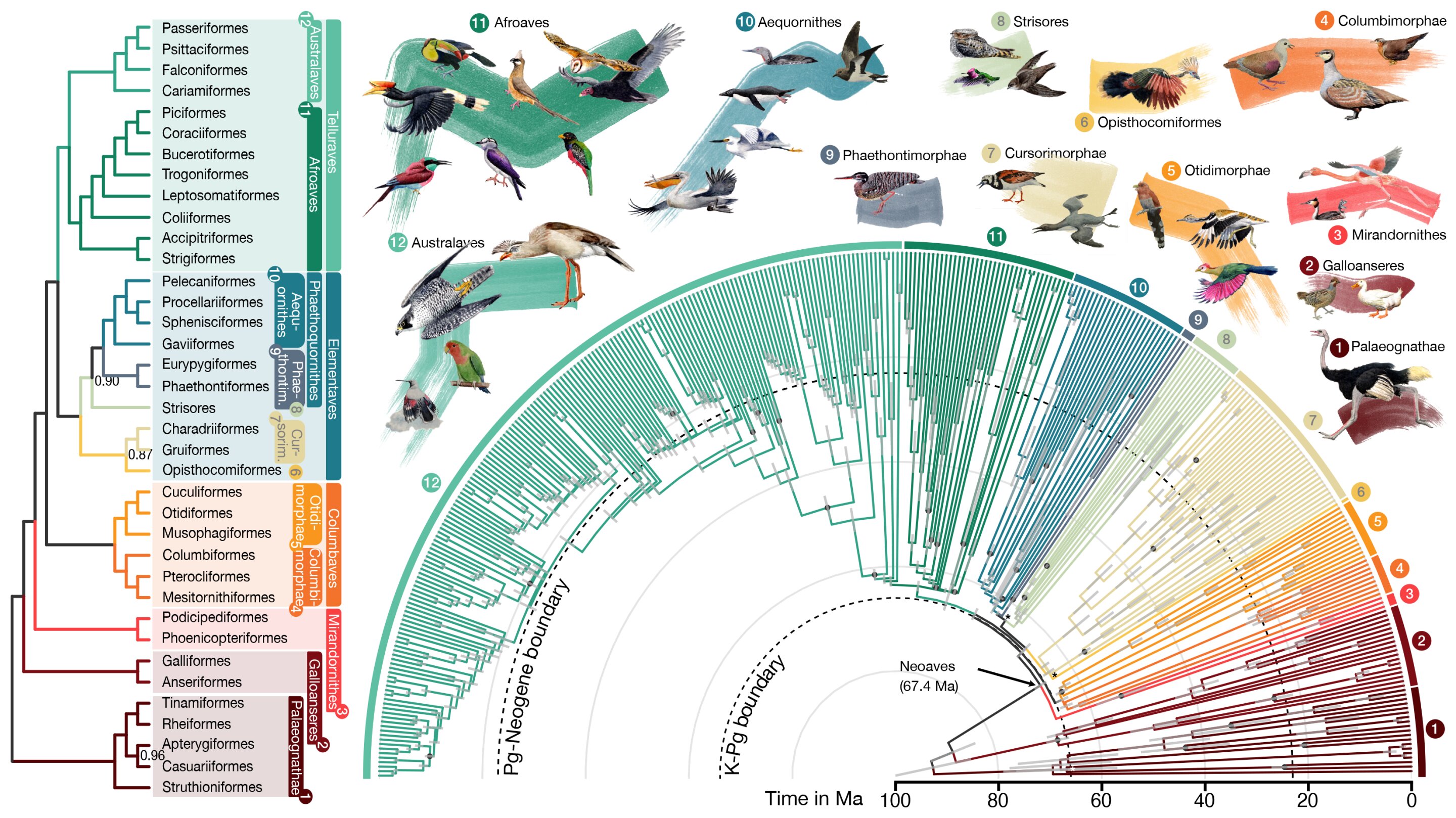
An international team of scientists has reconstructed the largest and most detailed bird family tree to date, delineating 93 million years of evolutionary relationships between 363 bird species using cutting-edge computational methods. The study, published in Nature and PNAS, revealed insights into avian diversification following the mass extinction event that wiped out the dinosaurs, and corrected previous misconceptions about the evolutionary relationships between bird species. The advanced computational tools developed by engineers at the University of California San Diego played a crucial role in analyzing vast amounts of genomic data with high accuracy and speed, paving the way for the construction of the most comprehensive bird family tree ever assembled.