"Meta AI Expands Drug Discovery Potential with Protein Structure Analysis"
TL;DR Summary
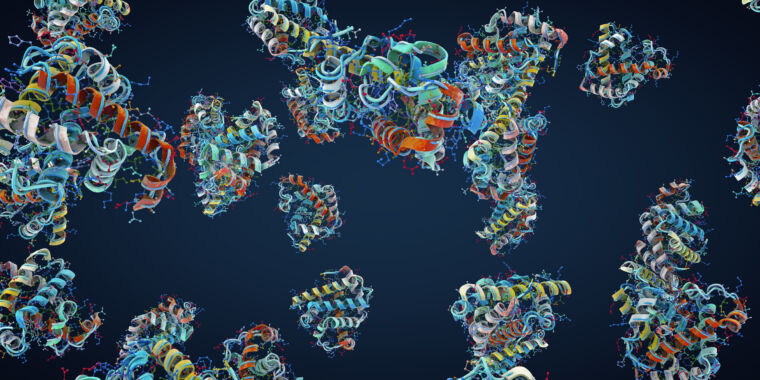
A team at Meta has trained a large language model (LLM) on the statistics of the appearance of amino acids within proteins and used the system's internal representation of what it learned to extract information about the structure of those proteins. The resulting system, ESM-2, was able to ingest a raw string of amino acids and output a 3D protein structure, along with a score that represents its confidence in the accuracy of that structure. This approach is faster than the best competing AI systems for predicting protein structures and still improving.
Topics:health#ai-systems#amino-acids#artificial-intelligence#large-language-models#neural-networks#protein-structures
- Large language models also work for protein structures Ars Technica
- Meta AI Unlocks Hundreds of Millions of Proteins to Aid Drug Discovery The Wall Street Journal
Reading Insights
Total Reads
0
Unique Readers
6
Time Saved
4 min
vs 5 min read
Condensed
90%
954 → 92 words
Want the full story? Read the original article
Read on Ars Technica