Unveiling Protein Stability, Design, and Recognition Mechanisms: Insights from Probabilistic Views and AI
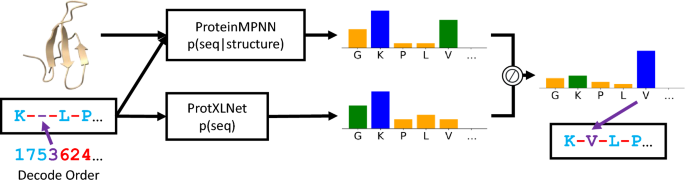
Researchers propose a new approach called BayesDesign for protein sequence design that maximizes the Boltzmann probability objective function \(p(\text {structure}|\text {seq})\) without relying on gradient descent or MCMC optimization techniques. The study mathematically formalizes protein design objectives for stability and conformational specificity and shows how they relate to the Boltzmann probability objective. The BayesDesign algorithm is evaluated on two model systems, NanoLuciferase enzyme and the WW beta sheet motif, and the designed sequences show increased stability and conformational specificity compared to the native sequences. The approach offers a faster and more reliable way to design proteins with desired properties.
- A probabilistic view of protein stability, conformational specificity, and design | Scientific Reports Nature.com
- Google’s AI protein folder IDs structure where none seemingly existed Ars Technica
- Combined prediction and design reveals the target recognition mechanism of an intrinsically disordered protein interaction domain | Proceedings of the National Academy of Sciences pnas.org
Reading Insights
0
5
25 min
vs 26 min read
98%
5,029 → 99 words
Want the full story? Read the original article
Read on Nature.com